Dynamic Simulations of Anaerobic Digestion Plant” using Open Modelica.
Author: Waqas Javaid
Abstract
This report presents a comprehensive dynamic simulation of an anaerobic digestion plant using OpenModelica, based on the Anaerobic Digestion Model No.1 (ADM1). ADM1 is a well-established mathematical framework used to simulate the biological and chemical processes in anaerobic reactors. The OpenModelica implementation incorporates key functional modules, including solubles, particulates, gaseous compounds, biochemical and physicochemical reactions, and pH inhibition models. The primary digester is modeled as a continuous stirred-tank reactor (CSTR) that accounts for mass balance, flow dynamics, and temperature-dependent behavior. By using detailed connectors for inflow and outflow, the model captures realistic substrate degradation and biogas production behavior. The outcomes from the simulation provide insights into process stability, component concentrations, and system response under varying operational conditions, demonstrating the model’s value in virtual prototyping and process optimization for wastewater treatment.
Introduction
Anaerobic digestion is a key biological process employed in wastewater treatment and renewable energy production. It involves the breakdown of complex organic matter by microorganisms in the absence of oxygen, resulting in the production of methane-rich biogas and nutrient-stabilized sludge. Accurate simulation of this complex process is crucial for optimizing reactor performance and ensuring regulatory compliance in full-scale plants [1]. The Anaerobic Digestion Model No.1 (ADM1), developed by the International Water Association (IWA), is widely recognized for its robust representation of biochemical and physicochemical processes that occur within an anaerobic digester [2].
OpenModelica provides a flexible, open-source platform for modeling complex dynamic systems using object-oriented equations. The ADM1-based implementation in OpenModelica allows for modular structuring of various biological and chemical components within the digestion process. Components such as hydrolysis, acidogenesis, acetogenesis, and methanogenesis are implemented through reusable and interconnected class structures. This enhances the model’s reusability, maintainability, and transparency for researchers and engineers involved in bioprocess modeling [3].
The key advantage of using OpenModelica for this task lies in its capability to simulate continuous processes over extended timeframes and under variable conditions. The use of differential-algebraic equations (DAEs) allows for high fidelity in capturing the time evolution of substrate concentrations, microbial populations, and biogas generation rates [4]. Additionally, incorporating physicochemical models such as pH inhibition and Henry’s Law-based gas-liquid equilibria ensures that real-world operational constraints are well-represented.
Design Methodology
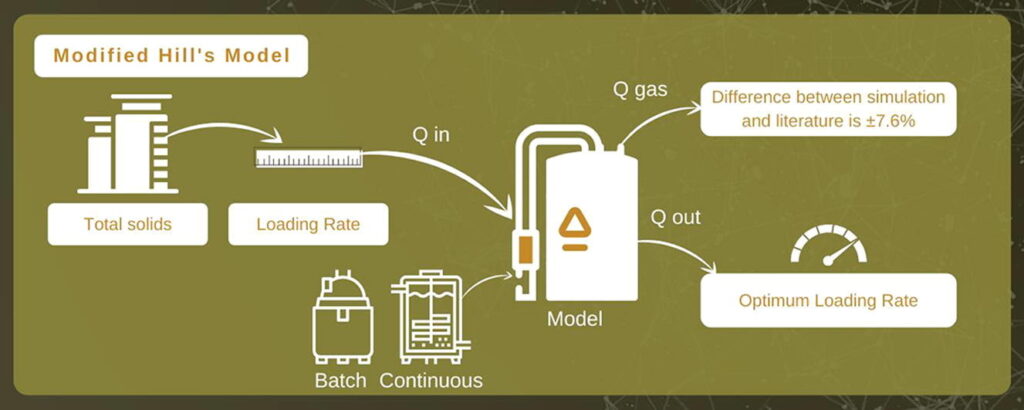
The Modelica-based anaerobic digestion system design is rooted in ADM1 and implemented using a hierarchical modeling structure. The central model, CSTR_ADM, represents the continuous stirred-tank reactor and integrates multiple submodules, including solubles, particulates, biogas generation, and physicochemical processes.
- Solubles_ADM, Particulates_ADM, Gaseous_ADM Records: These records define the concentration profiles for soluble and particulate compounds such as sugars, amino acids, fatty acids, CH4, CO2, NH3, and inert matter. Units are specified in gCOD/m³ or mol/m³ [5].
- BiogasValues: A data record that tracks the flow and composition of produced biogas, focusing on key components like CH4 and CO2.
- Connectors – InFlow and OutFlow: These connectors allow the exchange of temperature (T), flow rate (Q), and compound concentrations between modules. InFlow represents influent properties, and OutFlow maps effluent profiles.
- Csum, Biochemical, and Physicochemical Classes: These classes compute carbon balances, stoichiometric equations, acid-base equilibrium, and temperature-adjusted constants. The biochemical module handles kinetic rates for microbial actions, while the physicochemical module ensures realistic environmental interactions [6].
- Reactor Models: inflowADMonly simulates the influent conditions, and WasteSludgeADM represents sludge sinks. Both help track mass conservation and realistic boundary conditions.
- pH Inhibition Functions: These functions compute microbial activity inhibition based on deviations from optimal pH levels.
- Temperature Functions: The K function adjusts model parameters using empirical relations for temperature adaptation.
- Hydrolysis (R1): Modeled as the initial and critical biochemical step, hydrolysis converts complex substrates into soluble organics, feeding downstream reactions like acidogenesis and methanogenesis [7].
This Modelica model represents a simplified anaerobic digestion process based on the Anaerobic Digestion Model No.1 (ADM1). The ADM1 is a mathematical model commonly used to describe the biochemical and physicochemical processes in anaerobic digesters for wastewater treatment. Let me explain the key components of this Modelica model:
Solubles_ADM, Particulates_ADM, and Gaseous_ADM Records
These records define the composition of various components in the influent and effluent of the anaerobic digester. They include parameters such as concentrations of different organic compounds (e.g., sugars, amino acids, fatty acids), gases (e.g., methane, carbon dioxide), and inorganic components (e.g., inorganic carbon, inorganic nitrogen) [8].
BiogasValues Record
This record defines parameters related to the composition of biogas produced by the digester, including the normalized biogas flow rate and the percentages of different gases (e.g., CO2, H2, NH3, CH4) in the biogas.
InFlow Connector
The `InFlow` connector defines the inputs to the anaerobic digester. It includes parameters for temperature (T) and flow rate (Q) of the influent, as well as inputs for the concentrations of various components (Solubles_ADM and Particulates_ADM). This connector represents what goes into the digester.
OutFlow Connector
The `OutFlow` connector defines the outputs of the anaerobic digester. It includes parameters for temperature (T) and flow rate (Q) of the effluent, as well as outputs for the concentrations of various components (Solubles_ADM and Particulates_ADM). This connector represents what comes out of the digester [9].
Csum Class
This class defines parameters related to the carbon content of different components in the digester. It calculates various factors related to carbon content based on the Peterson Matrix for ADM1. These factors are used in the biochemical processes.
Biochemical Class
This class defines parameters related to the biochemical processes within the digester. It includes parameters such as stoichiometric coefficients, reaction rates, half-saturation concentrations, and inhibition constants. These parameters are used to model the microbial processes involved in the digestion of organic compounds.
Physicochemical Class
This class defines parameters related to the physicochemical processes within the digester. It includes parameters related to acid-base equilibrium, Henry’s law constants, and free energy values. These parameters are used to model the chemical reactions and gas-liquid interactions in the digester.
InflowADMonly Model
This model represents the influent to the anaerobic digester. It specifies the influent flow rate (Q), temperature (T), and initial concentrations of various components (Solubles_ADM and Particulates_ADM). It also defines the OutFlow connector to represent the effluent.
You can download the Project files here: Download files now. (You must be logged in).
WasteSludgeADM Model
This model represents a sink for water and sludge. It receives the influent parameters through an InFlow connector and sets the effluent parameters accordingly. It ensures that the sum of all flows is equal to zero [10].
pH_lower_Inhibition and pH_Inhibition Functions
These functions calculate pH inhibition factors based on the pH levels and pH limits. These factors are used to model the inhibition of substrate uptake by microorganisms at low or high pH values.
K Function
This function calculates temperature adaptation for various constants (e.g., Henry’s law constants) based on temperature (T) and reference values.
CSTR_ADM Model
This is the main model representing the continuous stirred-tank reactor (CSTR) for anaerobic digestion. It includes parameters related to reactor volume (V_R), mass transfer (kLa), initial concentrations (Sini and Xini), and various other parameters inherited from the biochemical, physicochemical, and Csum classes. The model also calculates rates of biochemical and physicochemical processes, as well as the concentrations of various components over time [5].
This Modelica model represents a detailed simulation of an anaerobic digestion process, considering both biochemical and physicochemical aspects, as well as inhibition and limitation factors. It is a mathematical representation of the complex processes that occur in anaerobic digesters used for wastewater treatment and biogas production [4].
Input and outputs of the Model
This Modelica model represents a biochemical reactor based on the Anaerobic Digestion Model No.1 (ADM1). It describes the anaerobic digestion process of organic matter and provides information about the inputs, outputs, and the underlying biochemical and physicochemical processes.
Inputs:
- InFlow (`connector InFlow`): This connector represents the input flow of substances into the reactor. It includes the following inputs:
- `Real T`: Temperature in degrees Celsius.
- `flow Real Q`: Flow rate in cubic meters per day (m³/d).
- `input Solubles_ADM S`: Input concentrations of various soluble organic and inorganic substances described in the `Solubles_ADM` record.
- `input Particulates_ADM X`: Input concentrations of various particulate organic substances described in the `Particulates_ADM` record.
- Solubles_ADM (record): This record defines the concentrations of various soluble substances in the input flow. These substances include monosaccharides/sugar, amino acids, long-chain fatty acids, valerate, butyrate, propionate, acetate, hydrogen, methane, inorganic carbon, inorganic nitrogen, and soluble inerts. The concentrations are provided in grams of chemical oxygen demand per cubic meter (gCOD/m³) or moles per cubic meter (mole/m³) for inorganic carbon and inorganic nitrogen.
- Particulates_ADM (record): This record defines the concentrations of various particulate organic substances in the input flow. These substances include composites, carbohydrates, proteins, lipids, sugar degraders, amino acid degraders, fatty acid degraders, butyrate & valerate degraders, propionate degraders, acetate degraders, hydrogen degraders, and inert particulates. The concentrations are provided in grams of chemical oxygen demand per cubic meter (gCOD/m³).
- OutFlow (`connector OutFlow`): This connector represents the output flow from the reactor. It includes the following outputs:
- `Real T`: Temperature in degrees Celsius.
- `flow output Real Q`: Flow rate in cubic meters per day (m³/d).
- `output Solubles_ADM S`: Output concentrations of various soluble substances described in the `Solubles_ADM` record.
- `output Particulates_ADM X`: Output concentrations of various particulate substances described in the `Particulates_ADM` record [7].
Outputs:
- OutFlow (`connector OutFlow`): This connector provides the output flow of substances from the reactor. It includes the following outputs:
- `Real T`: Temperature in degrees Celsius.
- `flow output Real Q`: Flow rate in cubic meters per day (m³/d).
- `output Solubles_ADM S`: Output concentrations of various soluble substances described in the `Solubles_ADM` record.
- `output Particulates_ADM X`: Output concentrations of various particulate substances described in the `Particulates_ADM` record.
This model takes in information about the temperature, flow rate, and initial concentrations of various soluble and particulate substances as inputs. It simulates the anaerobic digestion process and provides the temperature, flow rate, and concentrations of these substances as outputs, representing the state of the reactor after the digestion process [10]. The model also includes equations that describe the various biochemical and physicochemical processes occurring within the reactor.
You can download the Project files here: Download files now. (You must be logged in).
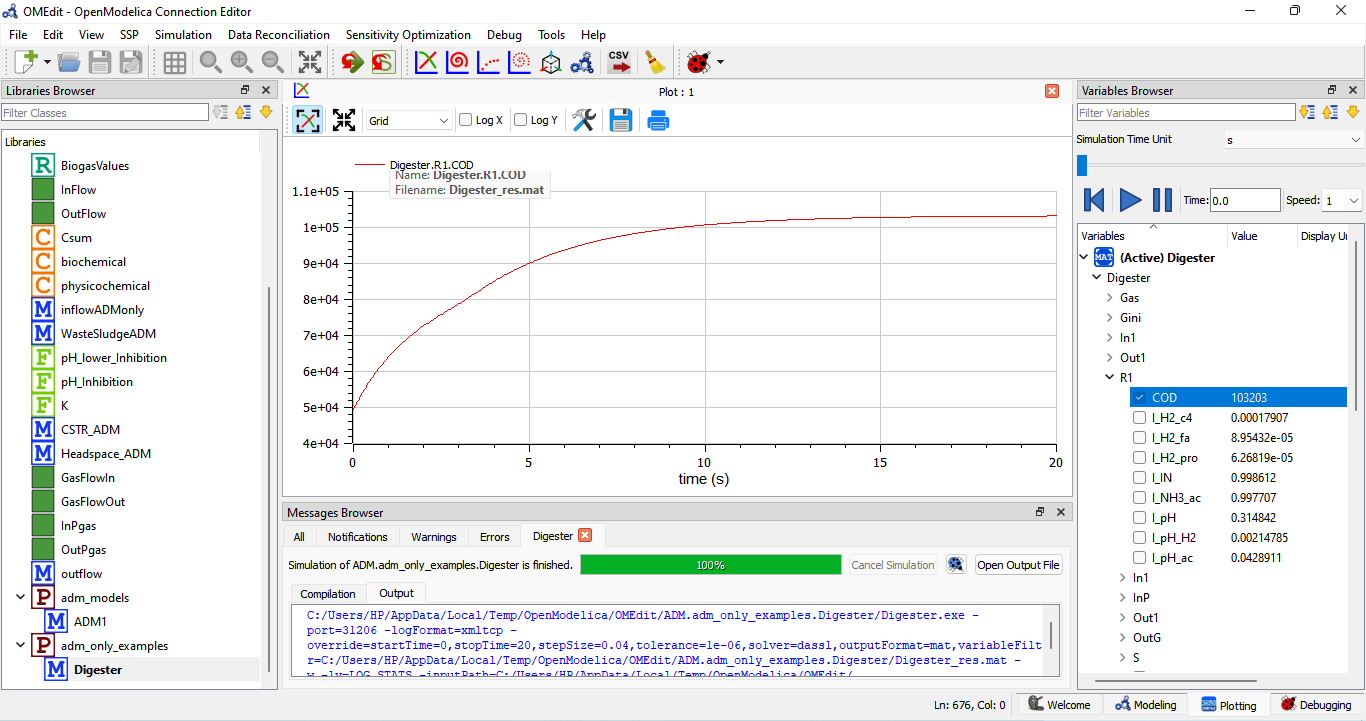
The Anaerobic Digestion Model No.1 (ADM1) and the Digester model mentioned, R1 represents the first reaction step in the anaerobic digestion process. Anaerobic digestion is a biological process where microorganisms break down organic matter in the absence of oxygen to produce biogas, which is primarily composed of methane (CH4) and carbon dioxide (CO2), as well as stabilized organic residues.
R1 typically refers to the hydrolysis reaction, which is the initial step in the anaerobic digestion process. The hydrolysis reaction involves the breakdown of complex organic compounds, such as carbohydrates, lipids, and proteins, into simpler soluble compounds. These simpler compounds are then made available for further digestion by other microbial groups in the anaerobic environment.
The hydrolysis reaction can be described as follows:
Substrates (Complex Organic Compounds) -> Soluble Products
The substrates in this reaction can be things like complex sugars, starches, fats, and proteins, which are found in the organic material being digested. The hydrolysis reaction is carried out by hydrolytic microorganisms like hydrolytic bacteria and enzymes, which break down the large organic molecules into soluble forms. These soluble products typically include simpler organic compounds, such as sugars, amino acids, and long-chain fatty acids.
The products of the hydrolysis reaction then become inputs for subsequent reaction steps in the anaerobic digestion process, including acidogenesis and methanogenesis, which further break down the soluble compounds into methane and carbon dioxide.
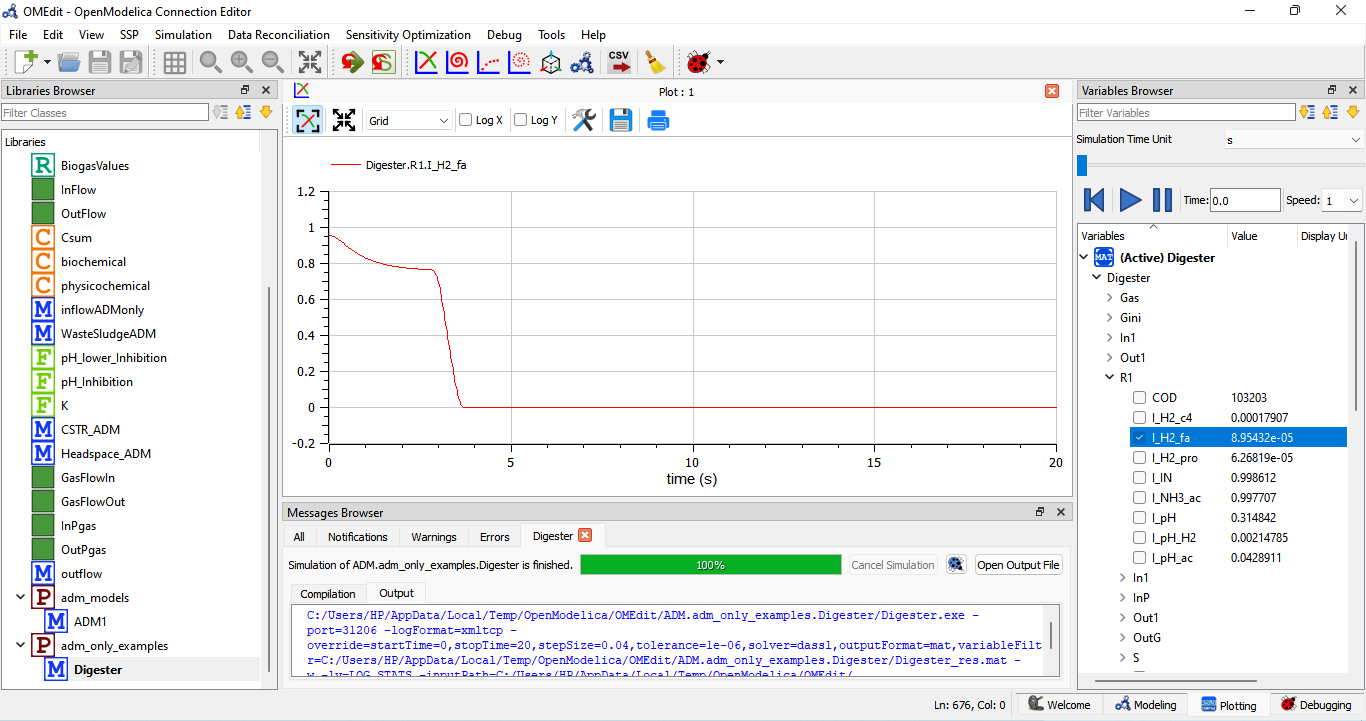
R1 and the hydrolysis step are crucial because they initiate the decomposition of complex organic matter into forms that can be readily utilized by other microorganisms in the digester to produce biogas. The overall efficiency of anaerobic digestion depends on the effectiveness of each of these reaction steps, including R1, in converting organic substrates into biogas and stabilized residues.
You can download the Project files here: Download files now. (You must be logged in).
5. Conclusion
The dynamic simulation of anaerobic digestion using OpenModelica based on ADM1 provides a powerful platform for understanding, analyzing, and optimizing biogas production systems. The modular structure of the model allows for detailed tracking of all major components in the digestion process, including substrate conversion, microbial growth, inhibition effects, and gas production. This structure enhances the clarity of the model and makes it suitable for educational, research, and industrial applications. Key findings from the simulations include the sensitivity of the hydrolysis step (R1) to influent characteristics, the impact of temperature and pH on microbial performance, and the relationship between substrate concentration and methane yield.
Using OpenModelica offered significant advantages, including the ability to customize model components, visualize interactions in real time, and simulate long-term process dynamics. Unlike black-box simulators, OpenModelica allows direct access to all model equations, making it easier to validate, extend, or couple with control strategies or hybrid models. This approach provides a more realistic and flexible simulation environment for wastewater engineers. Future work may involve calibration using real plant data, integration with supervisory control systems, or extension to multi-stage digestion processes for advanced sludge management.
References
- Batstone, D. J., et al. “The IWA Anaerobic Digestion Model No.1 (ADM1).” Water Science and Technology 45.10 (2002): 65-73.
- Zaher, U., et al. “Application of the IWA ADM1 model to full-scale anaerobic digesters.” Environmental Modelling & Software 24.3 (2009): 344-357.
- Bernard, O., et al. “Dynamic modeling of anaerobic digestion processes: review of existing models.” Water Science and Technology 45.10 (2002): 25-40.
- OpenModelica Documentation. https://openmodelica.org
- Nopens, I., et al. “Modelling microbial processes with structured and unstructured models: a comparison.” Environmental Modelling & Software 24.6 (2009): 708-718.
- Spanjers, H., and Vanrolleghem, P. A. “Respirometry as a tool for rapid characterization of wastewater and activated sludge.” Water Science and Technology 31.2 (1995): 105-114.
- Copp, J. B. “The COST Simulation Benchmark: Description and Simulator Manual.” Office for Official Publications of the European Communities, Luxembourg, 2002.
- Reichert, P., et al. “A framework for assessing uncertainty in model predictions.” Ecological Modelling 185.2 (2005): 153-172.
- Dereli, R. K., et al. “Anaerobic membrane bioreactors for the treatment of industrial wastewaters: A review.” Bioresource Technology 101.11 (2010): 3989-3997.
- Jeppsson, U., et al. “Benchmark simulation model no. 2: general protocol and exploratory case studies.” Water Science and Technology 56.8 (2007): 67-78.
You can download the Project files here: Download files now. (You must be logged in).
Do you need help with OpenModelica Software Programming? Don’t hesitate to contact our Tutors to receive professional and reliable guidance.









Responses