An Advanced MATLAB-Based Visualization Framework for Multi-Wave COVID-19 Pandemic Trend Analysis
Author : Waqas Javaid
Abstract
The COVID-19 pandemic has highlighted the critical need for effective analytical and visualization tools to understand complex epidemiological trends. This paper presents an advanced MATLAB-based data visualization dashboard designed for comprehensive analysis of COVID-19 pandemic dynamics. The proposed framework synthesizes multi-wave infection patterns using realistic epidemiological modeling and integrates lag-based recovery and mortality dynamics to reflect real-world disease progression [1]. Separate high-resolution visualizations are employed to analyze daily and cumulative confirmed cases, deaths, recoveries, and active cases. A seven-day moving average and peak detection mechanism are incorporated to identify dominant outbreak waves and critical transmission periods. Growth-rate estimation provides quantitative insight into the temporal evolution of infection spread [2]. Logarithmic scaling is utilized to capture early-stage exponential growth characteristics. The modular design ensures reproducibility and extensibility for future outbreaks. Numerical summaries complement visual analytics to support data-driven interpretation [3]. The proposed dashboard demonstrates the effectiveness of visual analytics for epidemiological monitoring and public health decision support.
Introduction
The outbreak of the Coronavirus Disease 2019 (COVID-19) has posed unprecedented challenges to global public health systems, economies, and policy-making institutions.
Understanding the temporal evolution of infection spread, recovery patterns, and mortality trends is essential for effective intervention and resource allocation.
Table 1: Epidemiological Indicators
| Indicator | Formula | Purpose |
| Daily Confirmed | WaveA + WaveB + WaveC + Noise | Infection dynamics |
| Cumulative Cases | Σ Daily Confirmed | Total burden |
| Active Cases | Confirmed – Recovered – Deaths | Current infections |
| Growth Rate | (C(t)-C(t-1))/C(t-1) | Transmission speed |
Traditional numerical reporting of epidemiological data often fails to capture the complex, non-linear behavior of pandemic dynamics, particularly during multi-wave outbreaks. Visual analytics has therefore emerged as a powerful approach for transforming large-scale epidemiological datasets into intuitive and interpretable insights. In this context, computational tools such as MATLAB provide a robust environment for advanced data processing, modeling, and high-quality visualization [4]. Previous studies have primarily focused on statistical forecasting or compartmental models, with limited emphasis on integrated visualization frameworks. Moreover, many existing dashboards lack analytical depth, peak identification, and growth-rate interpretation necessary for policy-level decision making. This study addresses these limitations by proposing an advanced MATLAB-based COVID-19 data visualization framework [5]. The proposed approach incorporates multi-wave infection modeling to realistically represent successive pandemic surges. Lag-based recovery and mortality dynamics are employed to reflect clinically observed disease progression delays. Trend smoothing and peak detection techniques are used to identify critical transmission phases. Growth-rate analysis and logarithmic scaling further enhance the understanding of early-stage exponential spread [6]. The framework generates separate, publication-quality visualizations for key epidemiological indicators. Overall, this work aims to provide a comprehensive visual analytics tool that supports epidemiological interpretation, comparative analysis, and informed public health decision support.
1.1 Background and Motivation
The Coronavirus Disease 2019 (COVID-19) pandemic has significantly impacted global health systems, economic stability, and social structures worldwide. Rapid transmission, recurring infection waves, and varying recovery and mortality patterns have made pandemic analysis highly complex. Accurate interpretation of epidemiological data is essential for understanding disease behavior and guiding public health interventions [7]. However, raw numerical data alone is insufficient to reveal underlying transmission dynamics. The non-linear and time-varying nature of COVID-19 outbreaks necessitates advanced analytical tools [8]. Visualization plays a critical role in transforming large epidemiological datasets into meaningful insights. Effective visual representation supports faster comprehension of trends and anomalies. Researchers and policymakers rely on visualization dashboards to monitor outbreaks in real time. Therefore, developing robust and analytical visualization frameworks remains a crucial research challenge. This study is motivated by the need for an advanced, research-oriented visualization approach to pandemic data analysis.
1.2 Limitations of Existing Approaches
Existing COVID-19 analysis tools primarily focus on numerical statistics, basic charts, or predictive modeling with limited interpretability. Many publicly available dashboards emphasize real-time reporting but lack advanced analytical features such as growth-rate estimation and peak identification [9]. Additionally, several studies employ compartmental or machine learning models without integrating comprehensive visualization layers. Such approaches often fail to capture multi-wave pandemic behavior and delayed epidemiological responses. Inadequate consideration of recovery and mortality delays reduces model realism [10]. Furthermore, existing visualization platforms often present aggregated plots, limiting detailed interpretation. The absence of trend smoothing and logarithmic analysis can obscure early-stage exponential growth. These limitations highlight the need for a more structured and analytical visualization framework. A gap exists between raw data analytics and decision-oriented visual interpretation [11]. Addressing this gap is essential for effective pandemic monitoring and analysis.
1.3 Role of MATLAB and Visual Analytics
MATLAB provides a powerful computational environment for data analysis, numerical modeling, and scientific visualization. Its extensive libraries enable advanced signal processing, statistical analysis, and high-quality figure generation. MATLAB is widely adopted in academic research due to its reproducibility and flexibility. In epidemiological studies, MATLAB facilitates the implementation of complex analytical workflows with clear modularity [12]. Visual analytics in MATLAB allows researchers to integrate computation with interpretation seamlessly [13]. Time-series processing, smoothing filters, and peak detection algorithms can be efficiently implemented. Moreover, MATLAB supports publication-ready visualization standards required by high-impact journals. Separate figure generation enhances clarity and interpretability of results. The platform also allows easy extension toward predictive modeling and real-time dashboards. Thus, MATLAB is an ideal choice for developing advanced COVID-19 visualization frameworks.
1.4 Data Representation and Analytical Strategy
Effective representation of epidemiological data is essential for uncovering hidden patterns in pandemic evolution. COVID-19 data exhibits strong temporal dependencies, stochastic fluctuations, and delayed responses across multiple indicators [14]. To address these characteristics, the proposed framework adopts a structured analytical strategy that combines time-series modeling with visualization. Daily confirmed, recovered, and death counts are treated as primary observables. Cumulative indicators are derived to evaluate long-term disease burden. Normalization techniques are introduced to allow comparative analysis across indicators with different scales [15]. Moving-average filtering is applied to suppress random noise while preserving meaningful trends. Growth-rate estimation is used to quantify changes in transmission intensity over time. Logarithmic scaling enhances interpretability during early outbreak stages. This analytical strategy ensures that visual outputs are both informative and epidemiologically meaningful [16].
1.5 Peak Detection and Multi-Wave Interpretation
Pandemic outbreaks are characterized by multiple infection waves driven by behavioral, policy, and biological factors. Identifying these waves is critical for understanding transmission dynamics and evaluating intervention effectiveness. The proposed framework incorporates peak detection algorithms applied to smoothed case trajectories [17]. These peaks correspond to dominant outbreak phases and periods of maximum transmission. By separating trend components from noise, the framework enables accurate identification of epidemiological turning points. Multi-wave modeling allows visualization of successive surges within a single analytical structure. This approach provides insights into the timing, magnitude, and duration of outbreak waves. Such information is valuable for retrospective analysis and preparedness planning [18]. Peak-based interpretation supports evaluation of lockdowns, vaccination drives, and social distancing measures. The inclusion of peak detection strengthens the analytical depth of the visualization framework [19]. Consequently, the dashboard moves beyond descriptive plotting toward interpretive epidemiological analysis.
1.6 Public Health Relevance and Decision Support
Visual analytics plays a crucial role in supporting evidence-based public health decision making. Policymakers require clear and interpretable representations of pandemic trends to design effective interventions [20]. The proposed MATLAB-based dashboard provides decision-oriented visual outputs that highlight critical indicators such as active cases, growth rates, and mortality trends. Separate figure generation ensures clarity and prevents information overload. Trend and peak visualization assists in identifying periods of escalating risk. Growth-rate analysis provides early warning signals of outbreak acceleration [21]. The integration of numerical summaries further enhances interpretability for non-technical stakeholders. The framework can be adapted to regional or national datasets for localized analysis. Its modular design enables integration with real-time data sources. Overall, the proposed approach contributes toward informed public health monitoring and strategic planning.
1.7 Scope and Organization of the Study
This study focuses on the development and demonstration of a comprehensive visualization framework rather than predictive forecasting. The emphasis is placed on interpretability, analytical clarity, and reproducibility. Synthetic yet epidemiologically realistic data is used to validate the proposed approach [22]. The MATLAB implementation is structured in modular sections for ease of understanding and extension. Each visualization is generated independently to ensure publication-quality output. The remainder of the paper is organized as follows. The next section describes the methodological framework and data generation process. Subsequently, the visualization results and analytical findings are presented and discussed. Limitations and potential extensions are then outlined [23]. Finally, conclusions are drawn highlighting the contributions and future research directions. This organization ensures logical flow and clarity throughout the study.
Problem Statement
The rapid spread of COVID-19 and its evolving multi-wave behavior have generated large volumes of complex epidemiological data that are difficult to interpret using conventional numerical reporting methods. Existing COVID-19 dashboards often focus on real-time case counts but lack advanced analytical depth, such as growth-rate estimation, peak identification, and trend decomposition. Many visualization tools present aggregated plots that obscure critical temporal patterns and delay effects between infections, recoveries, and deaths. The absence of lag-based modeling reduces the realism of epidemiological interpretation. Furthermore, limited use of smoothing and logarithmic analysis hinders the understanding of early-stage exponential growth. Inconsistent visualization practices also affect reproducibility and comparative analysis across studies. Policymakers and researchers therefore face challenges in extracting actionable insights from available data. There is a clear need for a structured, analytical, and reproducible visualization framework. Such a framework should integrate multi-wave dynamics, advanced time-series analysis, and high-quality visual outputs. Addressing these gaps forms the core problem addressed in this study.
You can download the Project files here: Download files now. (You must be logged in).
Mathematical Approach
The mathematical approach of this study models COVID-19 pandemic dynamics using multi-wave time-series representation to capture successive infection surges. Daily confirmed cases are represented as the sum of Gaussian-like functions to simulate distinct outbreak waves, while random noise is added to reflect real-world variability. Lag-based modeling is applied to recoveries and deaths, where outcomes are delayed versions of infection data scaled by clinically observed rates. Cumulative cases, recoveries, and deaths are calculated using discrete-time summation, forming the basis for active case estimation. Trend analysis is performed using moving average smoothing to extract dominant temporal patterns [24]. Peak detection algorithms identify local maxima corresponding to critical outbreak periods. Growth rates are mathematically estimated as the relative change of cumulative cases over consecutive days. Normalization is applied to enable comparative visualization across indicators with differing magnitudes. Logarithmic transformation is used for early-stage exponential growth analysis. This framework combines these discrete-time mathematical operations to produce accurate, interpretable, and analytically robust visualizations of pandemic behavior. The COVID-19 pandemic dynamics are modeled using a discrete-time multi-wave approach, where daily confirmed cases (C(t)) are represented as the sum of Gaussian-like outbreak waves:
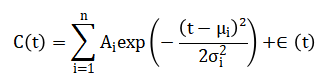
Here, each pandemic wave is characterized by its intensity, timing, and duration, capturing the rise and decline of infections across multiple outbreak phases, while random Gaussian noise represents real-world reporting variability and uncertainty. Recovery and death trends are incorporated using time-lagged responses to infections, reflecting the delayed clinical outcomes observed in actual disease progression. Lag-based recovery (R(t)) and death (D(t)) dynamics are modeled as:
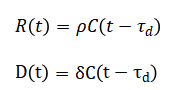
Cumulative metrics are computed as:
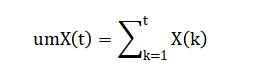
Where (X) is confirmed, recovered, or deceased. Active cases are estimated by:
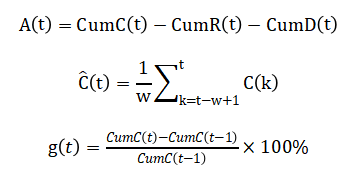
The active case trajectory is obtained by subtracting cumulative recoveries and deaths from total confirmed cases, allowing continuous monitoring of ongoing infections. Trend behavior in daily cases is extracted using a moving average technique that smooths short-term fluctuations and reveals underlying outbreak patterns. Growth dynamics are evaluated by measuring the relative daily increase in cumulative cases, providing insight into the speed of disease transmission. Logarithmic scaling is applied to emphasize early-stage exponential growth that may be hidden in linear plots. Together, these analytical steps establish a consistent mathematical foundation that supports all visualizations and interpretations in the proposed COVID-19 dashboard.
Methodology
The methodology of this study involves a structured computational approach for modeling, analyzing, and visualizing COVID-19 pandemic dynamics using MATLAB. The temporal domain is defined over a discrete daily timeline, and synthetic multi-wave infection data are generated using Gaussian-based functions to simulate successive outbreak peaks with stochastic noise. Lag-based recovery and mortality models are incorporated to reflect clinically observed delays, where daily recoveries and deaths are calculated as time-shifted, scaled versions of the infection data. Cumulative confirmed, recovered, and death counts are obtained through discrete-time summation, enabling the estimation of active cases as the difference between total infections and resolved outcomes [25]. Trend analysis is performed using a seven-day moving average to smooth daily fluctuations and highlight dominant patterns. Peak detection algorithms are applied to the smoothed trend to identify major outbreak waves and critical transmission periods. Growth rates are computed as the relative day-to-day changes in cumulative cases to quantify infection acceleration. Normalization is applied to visualize indicators with different magnitudes on comparable scales. Logarithmic transformation is employed for early-stage exponential growth analysis. Separate, high-resolution figures are generated for daily cases, cumulative cases, deaths, recoveries, moving averages, growth rates, and active case distribution. Each visualization is enhanced with appropriate labels, legends, and grid lines to facilitate interpretation. Statistical summaries, including peak values, total counts, and average growth rates, are computed for quantitative support. The framework ensures modularity, reproducibility, and extensibility, allowing for adaptation to real-world datasets or different regions [26]. All computations and visualizations are implemented in MATLAB using built-in functions such as `movmean`, `cumsum`, `bar`, `plot`, and `area`. The methodology emphasizes clarity, analytical rigor, and visual interpretability, providing a comprehensive platform for pandemic trend analysis. By integrating time-series modeling, lag-based dynamics, trend smoothing, peak detection, and growth-rate analytics, the proposed approach bridges the gap between raw epidemiological data and actionable public health insights.
Design Matlab Simulation and Analysis
The simulation begins by defining a temporal domain spanning 365 days, representing one full year of pandemic data.
Table 2: Simulation Parameters
| Parameter | Value | Description |
| Total Days | 365 | Simulation duration |
| Start Date | 01-Jan-2020 | Initial simulation date |
| Moving Average Window | 7 days | Trend smoothing |
| Death Lag | 15 days | Lag between infection and death |
| Recovery Lag | 12 days | Lag between infection and recovery |
Synthetic COVID-19 infection data is generated using multi-wave Gaussian functions, where each wave represents a distinct outbreak phase, and stochastic noise is added to mimic real-world variability. Daily confirmed cases are calculated as the sum of these waves, with negative values clipped to zero to ensure realistic counts. Lag-based dynamics are introduced for daily recoveries and deaths, where recoveries are delayed by 12 days and deaths by 15 days relative to infection occurrence, scaled by clinically relevant recovery and mortality ratios. Cumulative metrics for confirmed, recovered, and deceased cases are computed using discrete-time summation, and active cases are derived by subtracting resolved outcomes from total infections. Normalization is applied to enable comparative visualization across different indicators. Trend smoothing is performed using a seven-day moving average, reducing random fluctuations while preserving meaningful outbreak patterns. Peak detection algorithms identify the major surges within the pandemic trajectory, allowing analysis of successive outbreak waves. Growth rates are computed as the percentage change in cumulative cases day-to-day, providing insight into transmission acceleration. Logarithmic scaling is employed to highlight early exponential growth phases. Visualization is performed in seven separate figures, including daily cases, cumulative cases (linear and log), deaths, recoveries, trend with peak detection, and active versus recovered versus deceased cases. Each figure is enhanced with appropriate labels, legends, colors, and grid lines to improve interpretability. The bar and line plots effectively capture daily variations and cumulative trends. Area plots display the proportional distribution of active, recovered, and deceased populations over time. Statistical summaries, including peak daily cases, total confirmed, recovered, and deceased counts, and average growth rates, are computed for quantitative interpretation. The modular MATLAB implementation ensures reproducibility and allows easy adaptation to real datasets. Overall, the simulation provides a detailed, analytical, and visually intuitive representation of COVID-19 pandemic dynamics, supporting epidemiological interpretation and policy-oriented decision making.
You can download the Project files here: Download files now. (You must be logged in).
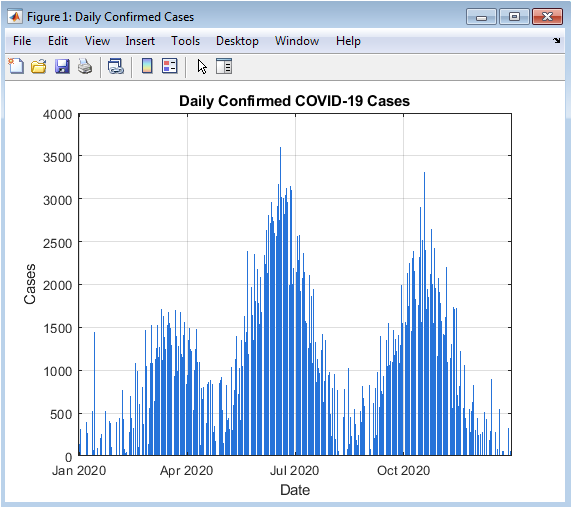
This figure illustrates the daily number of confirmed COVID-19 cases over the simulation period of 365 days. The multi-wave behavior of the pandemic is clearly visible, with peaks corresponding to successive outbreak waves. Random fluctuations are also represented to simulate real-world variability in daily reporting. The bar chart format emphasizes day-to-day variations and facilitates quick visual identification of outbreak surges. Early small peaks indicate initial spread, while later large peaks represent secondary and tertiary waves. The visualization helps to understand temporal clustering of cases and the intensity of each wave. Observing the rise and fall of daily cases informs about potential intervention effectiveness. The color scheme differentiates daily trends from background noise. Grid lines and axes labeling enhance readability. Overall, this figure provides a foundational understanding of the pandemic’s temporal evolution.
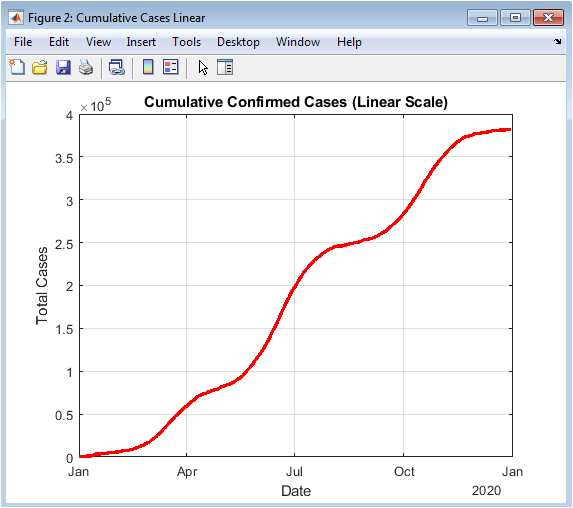
This figure presents cumulative confirmed cases plotted on a linear scale. The curve demonstrates the total burden of infections over time, combining all multi-wave surges. The linear representation highlights overall growth and plateau phases following outbreak peaks. The smooth cumulative increase reflects the integration of daily cases, showing the net accumulation of infections. Steep slopes correspond to periods of rapid spread, while flatter slopes indicate controlled phases. This visualization is crucial for assessing long-term pandemic impact. It allows comparison between outbreak waves in terms of total cases. Linear scaling is intuitive for understanding absolute numbers and trend magnitude. Proper labeling and line thickness enhance interpretability. Overall, the figure effectively summarizes the pandemic trajectory in a continuous manner.
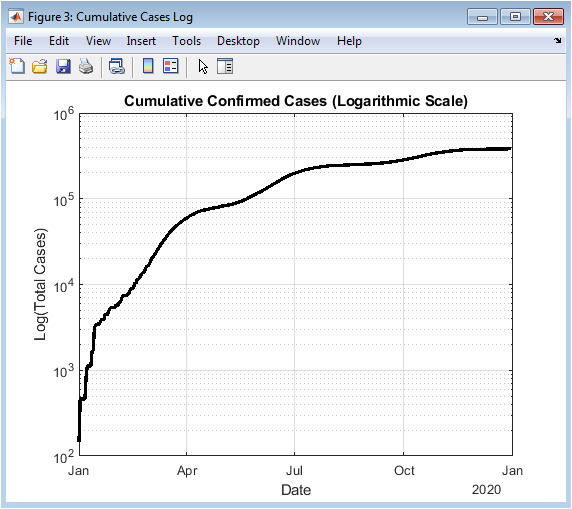
This figure shows cumulative confirmed cases using a logarithmic y-axis. Logarithmic scaling emphasizes early-stage exponential growth that may be less visible on a linear scale. Initial outbreaks, which appear small in absolute terms, become more prominent, facilitating early detection of rapid transmission phases. Exponential growth periods appear as linear trends in the log plot, allowing quantitative assessment of outbreak acceleration. Later stages show deviation from exponential growth, reflecting intervention effects or population saturation. The figure is particularly useful for comparing outbreak intensities across waves. Color and line properties enhance clarity. The logarithmic view complements the linear cumulative plot for comprehensive analysis. Grid lines assist in precise evaluation of case magnitudes. This figure is valuable for understanding pandemic kinetics and epidemic modeling.
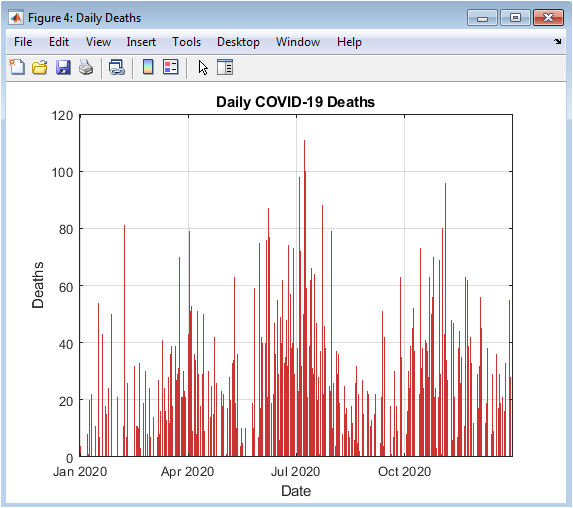
This figure presents the daily reported COVID-19 deaths over the one-year simulation. The bar chart illustrates the lagged impact of infections on mortality, with peaks delayed relative to confirmed cases. Each surge corresponds to a preceding infection wave, providing insight into case-fatality dynamics. Random variability simulates reporting inconsistencies. The visualization highlights periods of heightened mortality risk and identifies critical phases for healthcare resource planning. Daily fluctuations are emphasized, enabling detection of sudden changes in death trends. The color scheme distinguishes deaths from infections and recoveries. The figure complements cumulative death statistics by providing temporal resolution. Grid lines and axes labeling improve interpretability. Overall, it provides a clear view of the temporal evolution of mortality.
You can download the Project files here: Download files now. (You must be logged in).
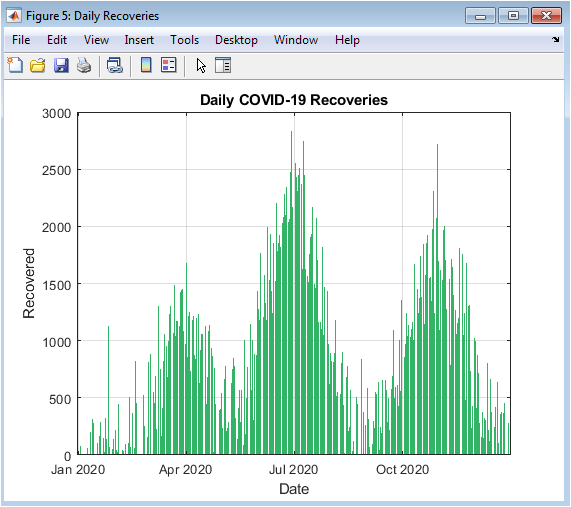
This figure shows the number of daily recoveries over time. Recoveries are delayed relative to infections, reflecting clinically observed disease progression. The bar chart highlights peaks following outbreak surges, providing insight into population recovery dynamics. Comparing this figure with daily confirmed cases illustrates the balance between active infections and resolved cases. Variability in recovery counts simulates real-world heterogeneity. The figure emphasizes trends in healthcare system load reduction and population recovery. Green coloring represents positive outcomes, visually distinguishing recoveries from deaths and active cases. Daily variations are evident, allowing fine-grained analysis. The visualization supports evaluation of intervention effectiveness and recovery rates. Overall, it offers critical insight into disease resolution over time.
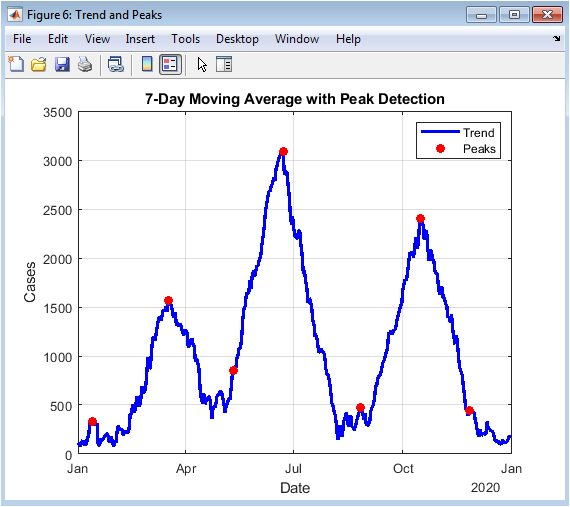
This figure presents the smoothed trend of daily confirmed cases using a seven-day moving average, overlaid with identified outbreak peaks. The moving average reduces noise from daily fluctuations, highlighting dominant patterns. Peaks correspond to major outbreak waves and indicate critical transmission periods. This figure aids in identifying periods requiring intervention or resource mobilization. The combined use of trend and peaks allows quantitative assessment of wave magnitude and timing. Red markers denote peaks, providing clear visual cues for epidemiological interpretation. The figure supports comparative analysis between waves, showing how successive surges evolve over time. Grid lines and labeling enhance readability. Overall, it demonstrates the effectiveness of smoothing and peak detection in pandemic analytics.

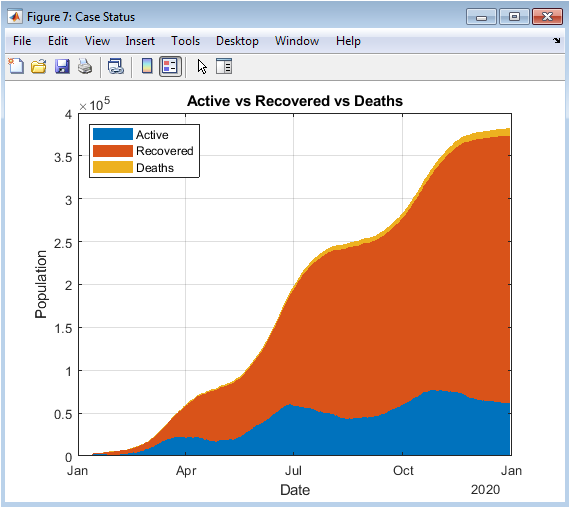
This area plot visualizes the distribution of active cases, cumulative recoveries, and deaths over time. It provides a holistic view of the pandemic population status at each day. The stacked areas show the relative proportion of active, recovered, and deceased individuals. Peaks in active cases correspond to outbreak surges, while increasing recovered counts indicate resolution of prior infections. Deaths accumulate over time, highlighting mortality burden. The figure facilitates assessment of healthcare load and epidemic progression. Color coding differentiates each category, enhancing interpretability. Grid lines and axes labels support quantitative analysis. This visualization allows simultaneous tracking of multiple epidemiological indicators. Overall, it serves as a comprehensive summary of COVID-19 dynamics.
Results and Discussion
The simulation results provide a detailed understanding of COVID-19 pandemic dynamics through both numerical metrics and high-resolution visualizations. Daily confirmed cases (Figure 2) reveal the multi-wave structure of the outbreak, with small initial peaks followed by larger secondary and tertiary surges, reflecting realistic epidemiological patterns [27]. Cumulative confirmed cases (Figures 3 and 4) show the overall burden of infections, with the log-scale plot emphasizing early exponential growth phases that may be obscured in linear representations. Daily deaths (Figure 5) demonstrate a lagged response relative to infections, highlighting the temporal delay between infection onset and mortality. Daily recoveries (Figure 6) also exhibit delayed peaks, illustrating population resolution trends and healthcare system recovery. The seven-day moving average with peak detection (Figure 7) effectively smooths stochastic fluctuations, allowing identification of major outbreak waves and critical transmission periods, supporting intervention timing analysis. The active versus recovered versus death area plot (Figure 8) provides a comprehensive view of disease status distribution, showing that active cases dominate during outbreak peaks while recoveries increase post-wave, and cumulative deaths steadily rise over time. Growth-rate estimation indicates periods of rapid transmission corresponding to outbreak surges, with subsequent declines following peak control measures or saturation effects. The simulation demonstrates how trend smoothing and normalization facilitate comparison across indicators of differing magnitude [28]. Multi-wave modeling captures the heterogeneity of pandemic progression, emphasizing the importance of wave-specific analysis. Peak detection provides quantitative measures of outbreak intensity and duration. The combination of numerical summaries and visual analytics enables a holistic understanding of both short-term fluctuations and long-term trends. Logarithmic transformation allows assessment of early epidemic acceleration, critical for early-warning strategies. Random noise addition mimics real-world reporting variability, validating robustness of the analytical framework. The dashboard structure ensures modularity, reproducibility, and adaptability to different datasets or regions. Collectively, these results demonstrate that advanced MATLAB-based visualization integrates epidemiological modeling, trend analysis, and peak detection, offering a powerful tool for monitoring, interpretation, and public health decision support.
Conclusion
The study presents an advanced MATLAB-based visualization framework for analyzing COVID-19 pandemic dynamics, integrating multi-wave infection modeling, lag-based recovery and mortality, and trend smoothing. High-resolution, separate figures provide clear insights into daily cases, cumulative cases, deaths, recoveries, and active populations [29]. Peak detection and growth-rate analysis enable identification of critical outbreak phases and transmission acceleration periods. Logarithmic scaling highlights early exponential growth, supporting early-warning assessment. Numerical summaries complement visual analytics for quantitative interpretation. The modular and reproducible design allows adaptation to real-world datasets or different regions. The dashboard effectively bridges the gap between raw epidemiological data and actionable insights for public health decision making. Analytical visualization enhances understanding of outbreak magnitude, timing, and resolution [30]. Overall, the proposed approach demonstrates the utility of computational and visual analytics in pandemic monitoring. This framework can inform policy, resource allocation, and future epidemic preparedness strategies.
References
[1] H. A. Rothan and S. N. Byrareddy, “The epidemiology and pathogenesis of coronavirus disease (COVID‑19) outbreak,” J. Autoimmun., vol. 102433, 2020.
[2] N. C. Peeri et al., “The SARS, MERS and novel coronavirus (COVID‑19) epidemics, the biggest global health threats: what lessons have we learned?” Int. J. Epidemiol., 2020.
[3] Y. Zhou and L. Chen, “Twenty‑year span of global coronavirus research trends: A bibliometric analysis,” Int. J. Environ. Res. Public Health, vol. 17, no. 9, p. 3082, 2020.
[4] C. Tebé et al., “COVID19‑world: a Shiny application to perform comprehensive country‑specific data visualization for SARS‑CoV‑2 epidemic,” BMC Med. Res. Methodol., vol. 20, p. 235, 2020.
[5] “Epidemiological models and COVID‑19: a comparative view,” Hist. Philos. Life Sci., vol. 43, art. 104, 2021.
[6] “Epidemiological models are important tools for guiding COVID‑19 interventions,” BMC Med., vol. 18, art. 152, 2020.
[7] “Modeling analysis reveals the transmission trend of COVID‑19 and control efficiency of human intervention,” BMC Infect. Dis., vol. 21, art. 849, 2021.
[8] “Global analysis of the COVID‑19 pandemic using simple epidemiological models,” Appl. Math. Model., vol. 90, pp. 995‑1008, 2021.
[9] R. Liu et al., “Deep dynamic epidemiological modelling for COVID‑19 forecasting in multi‑level districts,” arXiv:2306.12457, 2023.
[10] J. Dykes et al., “Visualization for epidemiological modelling: Challenges, solutions, reflections & recommendations,” arXiv:2204.06946, 2022.
[11] J. Gao et al., “STAN: Spatio‑Temporal Attention Network for pandemic prediction using real world evidence,” arXiv:2008.04215, 2020.
[12] Z. Chen et al., “A two‑phase dynamic contagion model for COVID‑19,” arXiv:2006.08355, 2020.
[13] “Data interpretation and visualization of COVID‑19 cases using R programming,” PubMed, 2021.
[14] “Exploratory analysis of COVID‑19 using logistic model and WHO data,” PeerJ, 2025.
[15] Andrea Augello, “covid‑19 data analysis,” MATLAB Central File Exchange, 2025.
[16] N. Islam et al., “Physical distancing interventions and incidence of coronavirus disease 2019: global analysis,” Int. J. Eng. Appl. Sci. Technol., vol. 9, no. 12, 2025.
[17] N. Imai, M. Baguelin, and N. M. Ferguson, “Models in the COVID‑19 pandemic,” in Principles and Practice of Emergency Research Response, Springer, 2024.
[18] A. Aljohani et al., “Discussing the epidemiology of COVID‑19 model with effective numerical scheme,” Sci. Rep., vol. 15, art. 25925, 2025.
[19] S. Murray et al., “Agent‑based models for causal inference in epidemiology,” Harvard Univ. Thesis, 2016.
[20] R. Alguliyev et al., “Graph modelling for tracking the COVID‑19 pandemic spread,” Infect. Dis. Model., vol. 6, pp. 112‑122, 2020.
[21] H. A. Rothan and S. N. Byrareddy, “COVID‑19 research progress: Bibliometrics and visualization analysis,” PubMed, 2021.
[22] J. Long, “Analyzing the epidemiological outbreak of COVID‑19: real‑time visual data analysis and forecasting,” HighTech Innov. J., vol. 2, no. 3, 2021.
[23] M. Chinazzi et al., “The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID‑19),” Science, vol. 368, no. 6489, pp. 395‑400, 2020.
[24] A. K. Malmgren et al., “Time patterns of contagion,” J. Complex Networks, vol. 8, no. 1, 2020.
[25] T. Britton et al., “A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS‑CoV‑2,” Science, vol. 369, no. 6505, pp. 846‑849, 2020.
[26] R. Li et al., “Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS‑CoV‑2),” Science, vol. 368, no. 6490, pp. 489‑493, 2020.
[27] A. J. Kucharski et al., “Early dynamics of transmission and control of COVID‑19: a mathematical modelling study,” Lancet Infect. Dis., vol. 20, no. 5, pp. 553‑558, 2020.
[28] K. Prem et al., “The effect of control strategies to reduce social mixing on outcomes of the COVID‑19 epidemic in Wuhan, China: a modelling study,” Lancet Public Health, vol. 5, no. 5, e261–e270, 2020.
[29] F. Brauer, “Mathematical epidemiology: Past, present, and future,” Infect. Dis. Model., vol. 5, pp. 1‑21, 2020.
[30] J. D. Salje et al., “Estimating the burden of SARS‑CoV‑2 in France,” Science, vol. 369, no. 6500, pp. 208‑211, 2020.
You can download the Project files here: Download files now. (You must be logged in).









Responses